Publication on local mutational processes in cancer genomes
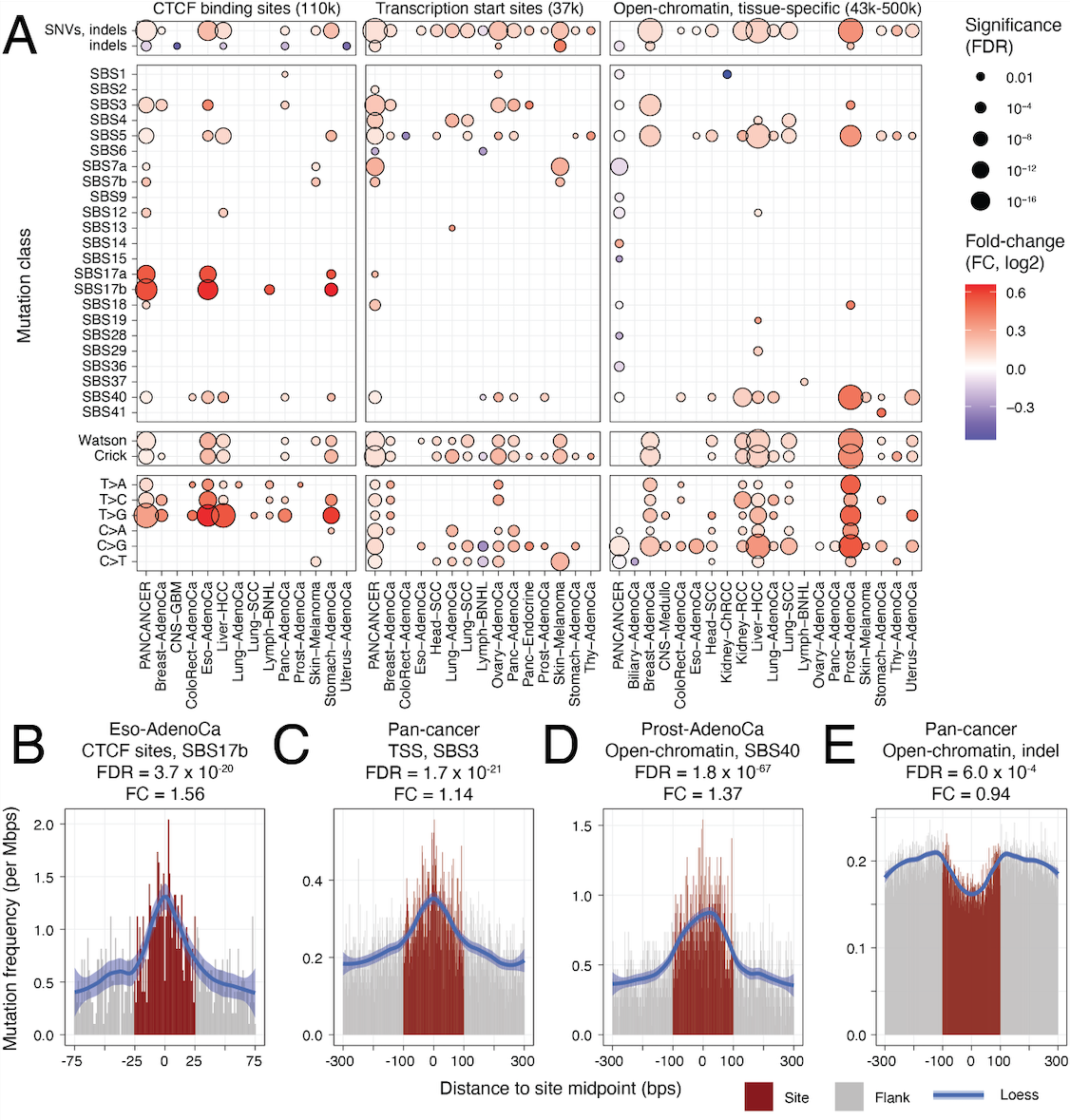
A new study on local mutational processes in cancer genomes is now published in Genome Biology. We studied the mutational processes affecting regulatory elements of the non-coding cancer genome involved in gene regulation and 3D chromatin architecture. We report novel functional and genetic correlates of local mutagenesis: transcription start sites (TSS) of active genes are highly enriched in mutations while TSSs of silent genes are not enriched; mutational processes are focused on constitutively active binding sites of CTCF that carry CTCF motifs, while sites bound in single or few tissues are not enriched in mutations; local mutagenic processes are particularly active in subsets of cancers with specific driver mutations, such as BRAF-mutant melanomas and ARID1A-mutant pancreatic cancers. We report a new open-source computational method, RM2, to analyze the mutational processes and signatures affecting entire classes of genomic elements using nucleotide-level, megabase-level and patient-level covariates. The study builds on the Pan-cancer Analysis of Whole Genomes (PCAWG) project and was co-led by Christian Lee, a graduate student of the Medical Biophysics Department of University of Toronto, and Diala Abd-Rabbo, a postdoctoral researcher of our lab.
See also: Tweet Thread and OICR News